 GelSim
GelSim

Gelsim - Students are given an unknown gene sequence, either circular or linear, to map. The DNA can be restricted with a range of enzymes, either singly or in pairs, and either completely or partially. Fragments can be cut out from gels and further restricted.
Here's an example of how it works.
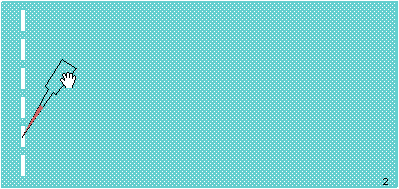
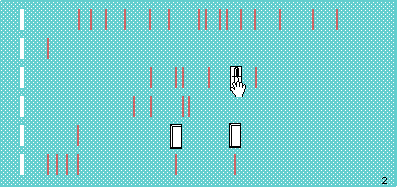
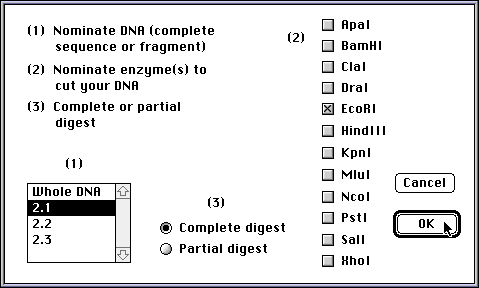
The student pours a gel, and then tells the program what restriction enzyme(s) to use. Here is the DNA about to be loaded into a slot:
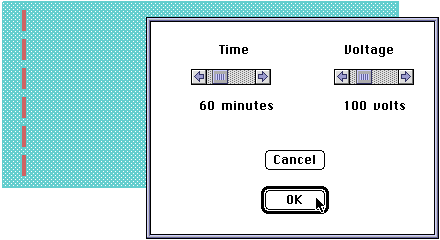
The gel is fully loaded, and ready to run.
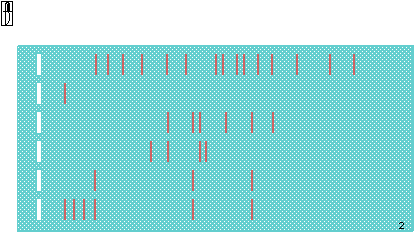
This is what it looks like after a run.
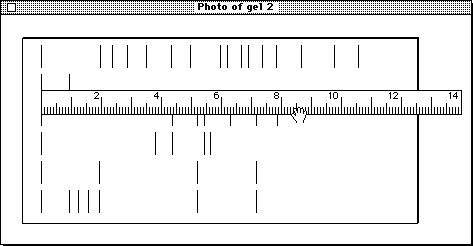
To measure the distances it's best to take a photo and work on that. There's a ruler to help, and unlike gels, photos can be stored.
Want to cut out some fragments for further restriction? That's what the knife is for.
Now when you go to nominate the digests for the next run, the cut out fragments are there.
The exercise of drawing up a map is not an easy one. To help understand the process, the program lets the student run a 'trial'. A new sequence is provided, and for this one the student can look up the map. Here's what a circular one looks like.

The map can also be checked with just one or two enzymes. That makes it easier to get a feeling for the relationship between the map and the fragment sizes. After running a trial with a known sequence, it is easier to go back and try the unknown sequence.